import pandas as pd
from sklearn.datasets import load_digits
digits = load_digits()Random Forest
Random Forest
Digits dataset from sklearn
dir(digits)['DESCR', 'data', 'feature_names', 'frame', 'images', 'target', 'target_names']import matplotlib.pyplot as pltplt.gray()
for i in range(4):
plt.matshow(digits.images[i])<Figure size 640x480 with 0 Axes>
df = pd.DataFrame(digits.data)
df.head()| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 0.0 | 5.0 | 13.0 | 9.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 6.0 | 13.0 | 10.0 | 0.0 | 0.0 | 0.0 |
| 1 | 0.0 | 0.0 | 0.0 | 12.0 | 13.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 11.0 | 16.0 | 10.0 | 0.0 | 0.0 |
| 2 | 0.0 | 0.0 | 0.0 | 4.0 | 15.0 | 12.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 11.0 | 16.0 | 9.0 | 0.0 |
| 3 | 0.0 | 0.0 | 7.0 | 15.0 | 13.0 | 1.0 | 0.0 | 0.0 | 0.0 | 8.0 | ... | 9.0 | 0.0 | 0.0 | 0.0 | 7.0 | 13.0 | 13.0 | 9.0 | 0.0 | 0.0 |
| 4 | 0.0 | 0.0 | 0.0 | 1.0 | 11.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 16.0 | 4.0 | 0.0 | 0.0 |
5 rows × 64 columns
df['target'] = digits.targetdf[0:12]| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 0.0 | 5.0 | 13.0 | 9.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 6.0 | 13.0 | 10.0 | 0.0 | 0.0 | 0.0 | 0 |
| 1 | 0.0 | 0.0 | 0.0 | 12.0 | 13.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 11.0 | 16.0 | 10.0 | 0.0 | 0.0 | 1 |
| 2 | 0.0 | 0.0 | 0.0 | 4.0 | 15.0 | 12.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 11.0 | 16.0 | 9.0 | 0.0 | 2 |
| 3 | 0.0 | 0.0 | 7.0 | 15.0 | 13.0 | 1.0 | 0.0 | 0.0 | 0.0 | 8.0 | ... | 0.0 | 0.0 | 0.0 | 7.0 | 13.0 | 13.0 | 9.0 | 0.0 | 0.0 | 3 |
| 4 | 0.0 | 0.0 | 0.0 | 1.0 | 11.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 16.0 | 4.0 | 0.0 | 0.0 | 4 |
| 5 | 0.0 | 0.0 | 12.0 | 10.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 9.0 | 16.0 | 16.0 | 10.0 | 0.0 | 0.0 | 5 |
| 6 | 0.0 | 0.0 | 0.0 | 12.0 | 13.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 1.0 | 9.0 | 15.0 | 11.0 | 3.0 | 0.0 | 6 |
| 7 | 0.0 | 0.0 | 7.0 | 8.0 | 13.0 | 16.0 | 15.0 | 1.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 13.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7 |
| 8 | 0.0 | 0.0 | 9.0 | 14.0 | 8.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 11.0 | 16.0 | 15.0 | 11.0 | 1.0 | 0.0 | 8 |
| 9 | 0.0 | 0.0 | 11.0 | 12.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | ... | 0.0 | 0.0 | 0.0 | 9.0 | 12.0 | 13.0 | 3.0 | 0.0 | 0.0 | 9 |
| 10 | 0.0 | 0.0 | 1.0 | 9.0 | 15.0 | 11.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 1.0 | 10.0 | 13.0 | 3.0 | 0.0 | 0.0 | 0 |
| 11 | 0.0 | 0.0 | 0.0 | 0.0 | 14.0 | 13.0 | 1.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 13.0 | 16.0 | 1.0 | 0.0 | 1 |
12 rows × 65 columns
Train and the model and prediction
X = df.drop('target',axis='columns')
y = df.targetfrom sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.2)from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(n_estimators=20)
model.fit(X_train, y_train)RandomForestClassifier(n_estimators=20)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
RandomForestClassifier(n_estimators=20)
?RandomForestClassifierInit signature: RandomForestClassifier( n_estimators=100, *, criterion='gini', max_depth=None, min_samples_split=2, min_samples_leaf=1, min_weight_fraction_leaf=0.0, max_features='sqrt', max_leaf_nodes=None, min_impurity_decrease=0.0, bootstrap=True, oob_score=False, n_jobs=None, random_state=None, verbose=0, warm_start=False, class_weight=None, ccp_alpha=0.0, max_samples=None, ) Docstring: A random forest classifier. A random forest is a meta estimator that fits a number of decision tree classifiers on various sub-samples of the dataset and uses averaging to improve the predictive accuracy and control over-fitting. The sub-sample size is controlled with the `max_samples` parameter if `bootstrap=True` (default), otherwise the whole dataset is used to build each tree. For a comparison between tree-based ensemble models see the example :ref:`sphx_glr_auto_examples_ensemble_plot_forest_hist_grad_boosting_comparison.py`. Read more in the :ref:`User Guide <forest>`. Parameters ---------- n_estimators : int, default=100 The number of trees in the forest. .. versionchanged:: 0.22 The default value of ``n_estimators`` changed from 10 to 100 in 0.22. criterion : {"gini", "entropy", "log_loss"}, default="gini" The function to measure the quality of a split. Supported criteria are "gini" for the Gini impurity and "log_loss" and "entropy" both for the Shannon information gain, see :ref:`tree_mathematical_formulation`. Note: This parameter is tree-specific. max_depth : int, default=None The maximum depth of the tree. If None, then nodes are expanded until all leaves are pure or until all leaves contain less than min_samples_split samples. min_samples_split : int or float, default=2 The minimum number of samples required to split an internal node: - If int, then consider `min_samples_split` as the minimum number. - If float, then `min_samples_split` is a fraction and `ceil(min_samples_split * n_samples)` are the minimum number of samples for each split. .. versionchanged:: 0.18 Added float values for fractions. min_samples_leaf : int or float, default=1 The minimum number of samples required to be at a leaf node. A split point at any depth will only be considered if it leaves at least ``min_samples_leaf`` training samples in each of the left and right branches. This may have the effect of smoothing the model, especially in regression. - If int, then consider `min_samples_leaf` as the minimum number. - If float, then `min_samples_leaf` is a fraction and `ceil(min_samples_leaf * n_samples)` are the minimum number of samples for each node. .. versionchanged:: 0.18 Added float values for fractions. min_weight_fraction_leaf : float, default=0.0 The minimum weighted fraction of the sum total of weights (of all the input samples) required to be at a leaf node. Samples have equal weight when sample_weight is not provided. max_features : {"sqrt", "log2", None}, int or float, default="sqrt" The number of features to consider when looking for the best split: - If int, then consider `max_features` features at each split. - If float, then `max_features` is a fraction and `max(1, int(max_features * n_features_in_))` features are considered at each split. - If "sqrt", then `max_features=sqrt(n_features)`. - If "log2", then `max_features=log2(n_features)`. - If None, then `max_features=n_features`. .. versionchanged:: 1.1 The default of `max_features` changed from `"auto"` to `"sqrt"`. Note: the search for a split does not stop until at least one valid partition of the node samples is found, even if it requires to effectively inspect more than ``max_features`` features. max_leaf_nodes : int, default=None Grow trees with ``max_leaf_nodes`` in best-first fashion. Best nodes are defined as relative reduction in impurity. If None then unlimited number of leaf nodes. min_impurity_decrease : float, default=0.0 A node will be split if this split induces a decrease of the impurity greater than or equal to this value. The weighted impurity decrease equation is the following:: N_t / N * (impurity - N_t_R / N_t * right_impurity - N_t_L / N_t * left_impurity) where ``N`` is the total number of samples, ``N_t`` is the number of samples at the current node, ``N_t_L`` is the number of samples in the left child, and ``N_t_R`` is the number of samples in the right child. ``N``, ``N_t``, ``N_t_R`` and ``N_t_L`` all refer to the weighted sum, if ``sample_weight`` is passed. .. versionadded:: 0.19 bootstrap : bool, default=True Whether bootstrap samples are used when building trees. If False, the whole dataset is used to build each tree. oob_score : bool or callable, default=False Whether to use out-of-bag samples to estimate the generalization score. By default, :func:`~sklearn.metrics.accuracy_score` is used. Provide a callable with signature `metric(y_true, y_pred)` to use a custom metric. Only available if `bootstrap=True`. n_jobs : int, default=None The number of jobs to run in parallel. :meth:`fit`, :meth:`predict`, :meth:`decision_path` and :meth:`apply` are all parallelized over the trees. ``None`` means 1 unless in a :obj:`joblib.parallel_backend` context. ``-1`` means using all processors. See :term:`Glossary <n_jobs>` for more details. random_state : int, RandomState instance or None, default=None Controls both the randomness of the bootstrapping of the samples used when building trees (if ``bootstrap=True``) and the sampling of the features to consider when looking for the best split at each node (if ``max_features < n_features``). See :term:`Glossary <random_state>` for details. verbose : int, default=0 Controls the verbosity when fitting and predicting. warm_start : bool, default=False When set to ``True``, reuse the solution of the previous call to fit and add more estimators to the ensemble, otherwise, just fit a whole new forest. See :term:`Glossary <warm_start>` and :ref:`gradient_boosting_warm_start` for details. class_weight : {"balanced", "balanced_subsample"}, dict or list of dicts, default=None Weights associated with classes in the form ``{class_label: weight}``. If not given, all classes are supposed to have weight one. For multi-output problems, a list of dicts can be provided in the same order as the columns of y. Note that for multioutput (including multilabel) weights should be defined for each class of every column in its own dict. For example, for four-class multilabel classification weights should be [{0: 1, 1: 1}, {0: 1, 1: 5}, {0: 1, 1: 1}, {0: 1, 1: 1}] instead of [{1:1}, {2:5}, {3:1}, {4:1}]. The "balanced" mode uses the values of y to automatically adjust weights inversely proportional to class frequencies in the input data as ``n_samples / (n_classes * np.bincount(y))`` The "balanced_subsample" mode is the same as "balanced" except that weights are computed based on the bootstrap sample for every tree grown. For multi-output, the weights of each column of y will be multiplied. Note that these weights will be multiplied with sample_weight (passed through the fit method) if sample_weight is specified. ccp_alpha : non-negative float, default=0.0 Complexity parameter used for Minimal Cost-Complexity Pruning. The subtree with the largest cost complexity that is smaller than ``ccp_alpha`` will be chosen. By default, no pruning is performed. See :ref:`minimal_cost_complexity_pruning` for details. .. versionadded:: 0.22 max_samples : int or float, default=None If bootstrap is True, the number of samples to draw from X to train each base estimator. - If None (default), then draw `X.shape[0]` samples. - If int, then draw `max_samples` samples. - If float, then draw `max(round(n_samples * max_samples), 1)` samples. Thus, `max_samples` should be in the interval `(0.0, 1.0]`. .. versionadded:: 0.22 Attributes ---------- estimator_ : :class:`~sklearn.tree.DecisionTreeClassifier` The child estimator template used to create the collection of fitted sub-estimators. .. versionadded:: 1.2 `base_estimator_` was renamed to `estimator_`. base_estimator_ : DecisionTreeClassifier The child estimator template used to create the collection of fitted sub-estimators. .. deprecated:: 1.2 `base_estimator_` is deprecated and will be removed in 1.4. Use `estimator_` instead. estimators_ : list of DecisionTreeClassifier The collection of fitted sub-estimators. classes_ : ndarray of shape (n_classes,) or a list of such arrays The classes labels (single output problem), or a list of arrays of class labels (multi-output problem). n_classes_ : int or list The number of classes (single output problem), or a list containing the number of classes for each output (multi-output problem). n_features_in_ : int Number of features seen during :term:`fit`. .. versionadded:: 0.24 feature_names_in_ : ndarray of shape (`n_features_in_`,) Names of features seen during :term:`fit`. Defined only when `X` has feature names that are all strings. .. versionadded:: 1.0 n_outputs_ : int The number of outputs when ``fit`` is performed. feature_importances_ : ndarray of shape (n_features,) The impurity-based feature importances. The higher, the more important the feature. The importance of a feature is computed as the (normalized) total reduction of the criterion brought by that feature. It is also known as the Gini importance. Warning: impurity-based feature importances can be misleading for high cardinality features (many unique values). See :func:`sklearn.inspection.permutation_importance` as an alternative. oob_score_ : float Score of the training dataset obtained using an out-of-bag estimate. This attribute exists only when ``oob_score`` is True. oob_decision_function_ : ndarray of shape (n_samples, n_classes) or (n_samples, n_classes, n_outputs) Decision function computed with out-of-bag estimate on the training set. If n_estimators is small it might be possible that a data point was never left out during the bootstrap. In this case, `oob_decision_function_` might contain NaN. This attribute exists only when ``oob_score`` is True. See Also -------- sklearn.tree.DecisionTreeClassifier : A decision tree classifier. sklearn.ensemble.ExtraTreesClassifier : Ensemble of extremely randomized tree classifiers. sklearn.ensemble.HistGradientBoostingClassifier : A Histogram-based Gradient Boosting Classification Tree, very fast for big datasets (n_samples >= 10_000). Notes ----- The default values for the parameters controlling the size of the trees (e.g. ``max_depth``, ``min_samples_leaf``, etc.) lead to fully grown and unpruned trees which can potentially be very large on some data sets. To reduce memory consumption, the complexity and size of the trees should be controlled by setting those parameter values. The features are always randomly permuted at each split. Therefore, the best found split may vary, even with the same training data, ``max_features=n_features`` and ``bootstrap=False``, if the improvement of the criterion is identical for several splits enumerated during the search of the best split. To obtain a deterministic behaviour during fitting, ``random_state`` has to be fixed. References ---------- .. [1] L. Breiman, "Random Forests", Machine Learning, 45(1), 5-32, 2001. Examples -------- >>> from sklearn.ensemble import RandomForestClassifier >>> from sklearn.datasets import make_classification >>> X, y = make_classification(n_samples=1000, n_features=4, ... n_informative=2, n_redundant=0, ... random_state=0, shuffle=False) >>> clf = RandomForestClassifier(max_depth=2, random_state=0) >>> clf.fit(X, y) RandomForestClassifier(...) >>> print(clf.predict([[0, 0, 0, 0]])) [1] File: ~/mambaforge/envs/cfast/lib/python3.11/site-packages/sklearn/ensemble/_forest.py Type: ABCMeta Subclasses:
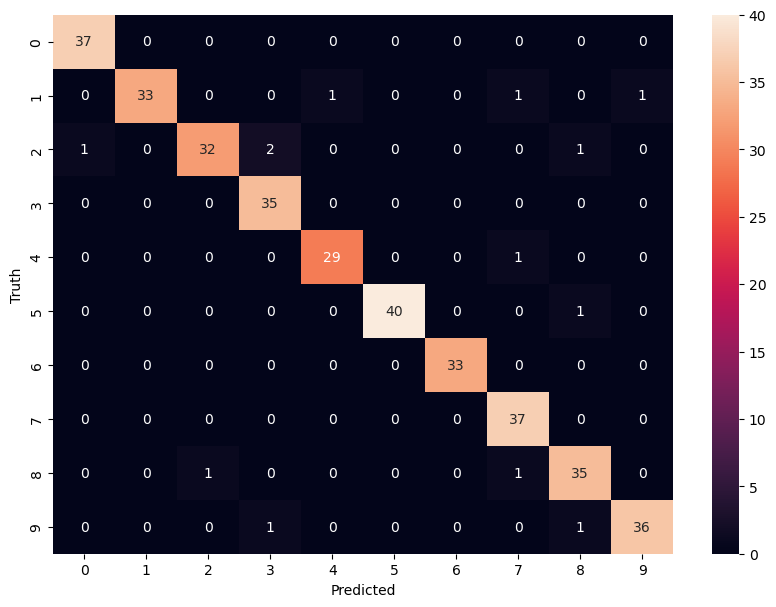
model.score(X_test, y_test)0.9638888888888889y_predicted = model.predict(X_test)Confusion Matrix
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_predicted)
cmarray([[37, 0, 0, 0, 0, 0, 0, 0, 0, 0],
[ 0, 33, 0, 0, 1, 0, 0, 1, 0, 1],
[ 1, 0, 32, 2, 0, 0, 0, 0, 1, 0],
[ 0, 0, 0, 35, 0, 0, 0, 0, 0, 0],
[ 0, 0, 0, 0, 29, 0, 0, 1, 0, 0],
[ 0, 0, 0, 0, 0, 40, 0, 0, 1, 0],
[ 0, 0, 0, 0, 0, 0, 33, 0, 0, 0],
[ 0, 0, 0, 0, 0, 0, 0, 37, 0, 0],
[ 0, 0, 1, 0, 0, 0, 0, 1, 35, 0],
[ 0, 0, 0, 1, 0, 0, 0, 0, 1, 36]])import matplotlib.pyplot as plt
import seaborn as sn
plt.figure(figsize=(10,7))
sn.heatmap(cm, annot=True)
plt.xlabel('Predicted')
plt.ylabel('Truth')Text(95.72222222222221, 0.5, 'Truth')